rheab
May 20, 2022, 1:41am
1
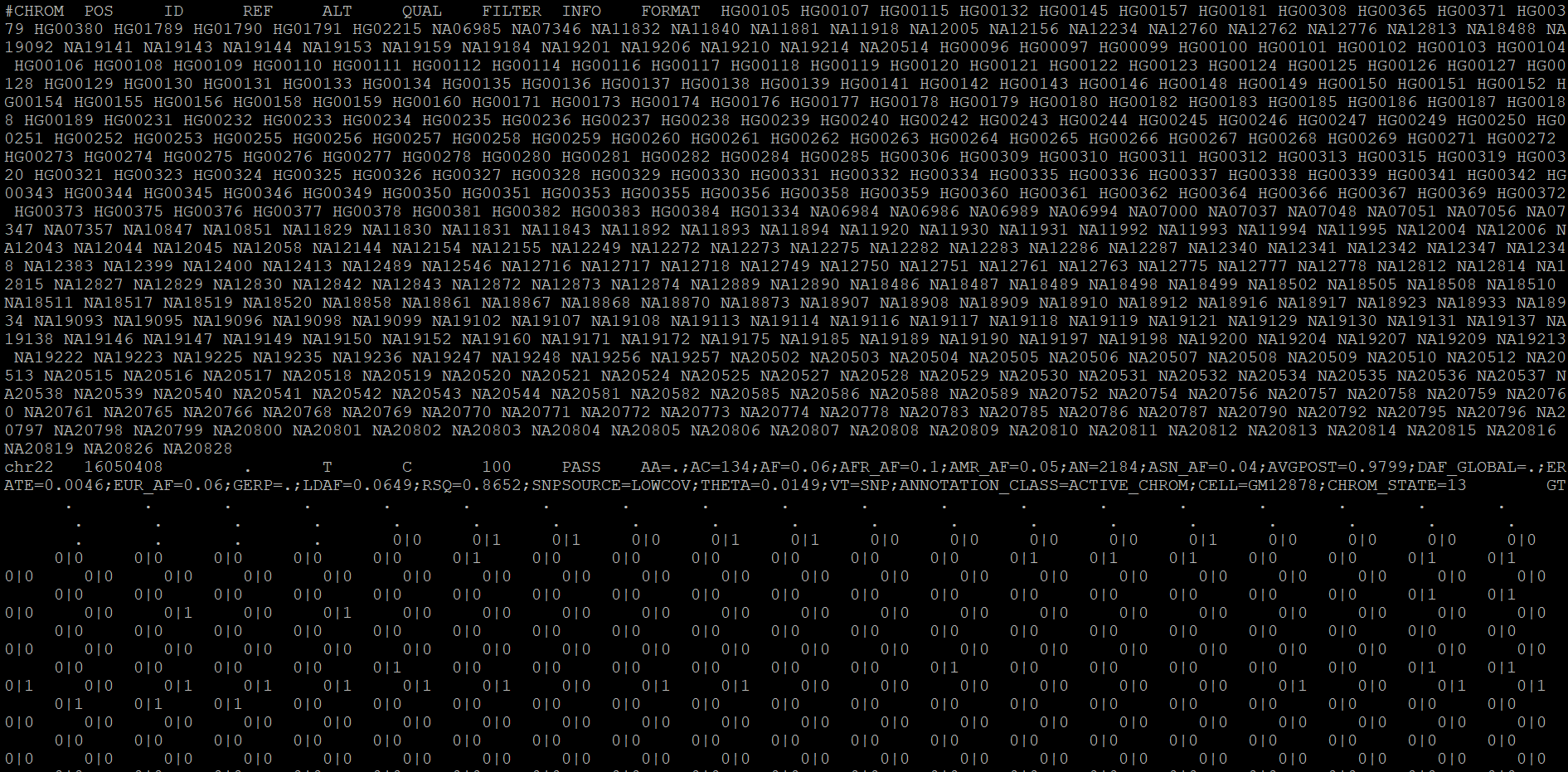
I have a vcf file. I need to update the ID column with this information.
0797 NA20798 NA20799 NA20800 NA20801 NA20802 NA20803 NA20804 NA20805 NA20806 NA20807 NA20808 NA20809 NA20810 NA20811 NA20812 NA20813 NA20814 NA20815 NA20816 NA20819 NA20826 NA20828
chr22 16050408 . T C 100.00 PASS AA=.;AC=134;AF=0.06;AFR_AF=0.10;AMR_AF=0.05;AN=2184;ASN_AF=0.04;AVGPOST=0.9799;DAF_GLOBAL=.;ERATE=0.0046;EUR_AF=0.06;GERP=.;LDAF=0.0649;RSQ=0.8652;SNPSOURCE=LOWCOV;THETA=0.0149;VT=SNP;ANNOTATION_CLASS=ACTIVE_CHROM;CELL=GM12878;CHROM_STATE=13
GT:GL:DS:PP:BD .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.:. .:.:.:.
So, I want to update this site "." with chr22_16050408_T_C_b37 so each ID column in my vcf file should look like chr{no.}position_refallele_altallele_b37.
I tried to use the following command but its now giving me the answer.
awk 'NR>1 {print $1""$2""$3""$4"_b37"}' genotype_chr22_filtered_dosage1.txt
@rheab , hi,
where is ID column in this ?
show ACTUAL EXPECTED output required.
use the MARKDOWN tags to display data not a stream of text that is hard to decipher please
is that single awk line all you attempted ?
TEAM, please wait till @rheab has responded with their attempt(s) before assisting
rheab
May 20, 2022, 3:25pm
3
I am able to remove the ID column using bcftools. Now I just need to update the ID column using the new format. I have editted my post.
@rheab , look at the response given in the link below.
However, we would appreciate if you can explain WHY this is being done (as we see a number of identical requests .... ).
I have a vcf file. I need to update the ID column in my vcf file. This is how my vcf file look like: [enter image description here]
0797 NA20798 NA20799 NA20800 NA20801 NA20802 NA20803 NA20804 NA20805 NA20806 NA20807 NA20808 NA20809 NA20810 NA20811 NA20812 NA20813 NA20814 NA20815 NA20816 NA20819 NA20826 NA20828
chr22 16050408 . T C 100 PASS AA=.;AC=134;AF=0.06;AFR_AF=0.1;AMR_AF=0.05;AN=2184;ASN_AF=0.04;AVGPOST=0.9799;DAF_GLOBAL=.;ERATE=0.0046;EUR_AF=0.06;GERP=.;LDAF=0.0649;RSQ=0.8652;SNPSOURCE…
Neo
May 24, 2022, 1:56am
6
This is either homework or multiple accounts asking the same question.